2 Tracks 3 Tasks
2 Tracks {CTA, MRA}
Participants can choose from two tracks, one track for CTA modality and one track for MRA modality. We encourage participants to take part in both tracks.

3 Tasks on CoW {Segmentation, Object Detection, Classification}
Task-1-CoW-Segmentation
The segmentation task is voxel-level multi-class CoW vessel segmentation, which is a repeated task continued from 2023 challenge.
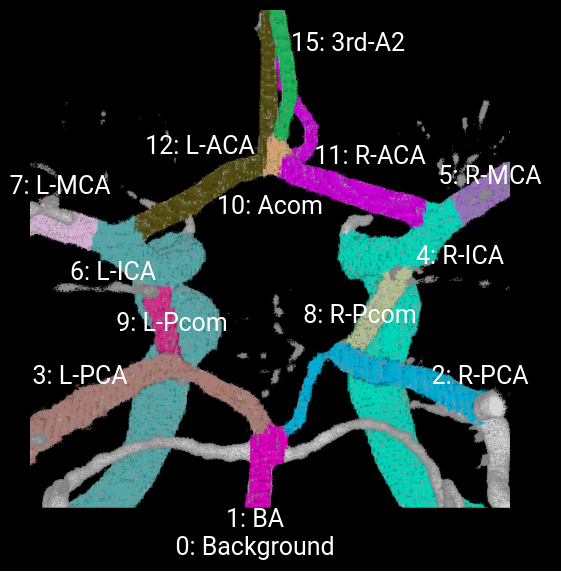
The vessel components of the CoW annotated are left and right internal carotid artery (ICA), left and right anterior cerebral artery (ACA), left and right middle cerebral artery (MCA), anterior communicating artery (Acom), left and right posterior communicating artery (Pcom), left and right posterior cerebral artery (PCA), and basilar artery (BA).
Here is the annotation mask in pixel value for each vessel objects of interest for a MRA example:
🎥 Videos of Task-1-CoW-Segmentation Labels
Example of a paired CTA (left) and MRA (right) with CoW segmentation mask annotations:


Note that the evaluations of segmentation results will be limited to within the CoW region of interest (ROI). (See below "Task-2-CoW-ObjDet" for more info on the CoW ROI.)
Task-2-CoW-ObjDet
The object detection ("objdet") task is to locate a 3D bounding box for the CoW region of interest (ROI).
The CoW ROI is defined as the 3D bounding box containing the volume required for the diagnosis of the CoW variant.

For each MRA or CTA image, we provide a text file containing the size and location of the 3D bounding box info.
- Size = number of pixels along the x, y, z axis
- Location = coordinate of the x-min, y-min, z-min (0-indexed)
An example bounding box txt file as released in roi_loc_labelsTr/topcow_ct_024.txt:
--- ROI Meta Data ---
Size (Voxels): 96 68 36
Location (Voxels): 103 93 77
Note that independent of whether taking part in Task-2-CoW-ObjDet, the evaluation of the Task-1-CoW-Segmentation results is limited to the CoW region of interest.
Task-3-CoW-Classification
The classification task is to classify the anterior and posterior parts of the CoW graph in the form of two edge lists per image.
For each image, the edges of the CoW graph are recorded in a yml file
indicating the presence of edges (0: absent, 1: present)
- 4 edges for
anteriorpart, roughly from left to right:- L-A1
- Acom
- 3rd-A2
- R-A1
- 4 edges for
posteriorpart, roughly from left to right:- L-Pcom
- L-P1
- R-P1
- R-Pcom
Here are three example cases to illustrate the classification annotation and task:
For example, the edge list for this CTA case is stored in the topcow_ct_145.yml file, and the edges are:

anterior:
L-A1: 1
Acom: 1
3rd-A2: 0
R-A1: 0
posterior:
L-Pcom: 0
L-P1: 1
R-P1: 1
R-Pcom: 0
The edges for this topcow_mr_163 case are listed as followed in topcow_mr_163.yml:

anterior:
L-A1: 1
Acom: 0
3rd-A2: 0
R-A1: 1
posterior:
L-Pcom: 1
L-P1: 1
R-P1: 1
R-Pcom: 1
The anterior and posterior edge lists, as annotated in topcow_mr_160.yml, for this topcow_mr_160 case are:

anterior:
L-A1: 1
Acom: 0
3rd-A2: 0
R-A1: 1
posterior:
L-Pcom: 1
L-P1: 0
R-P1: 1
R-Pcom: 1
